These are example input scripts, the full API can be found here.
A more in-depth tutorial with video recordings can be found here.
mumax3 myfile.mx3Output is automatically stored in the "myfile.out" directory. Additionally, a web interface provides live output. Default is
http://localhost:35367.mumax3 -help which will show the available command-line flags (e.g. to select a certain GPU).
SetGridsize(128, 32, 1) SetCellsize(500e-9/128, 125e-9/32, 3e-9) Msat = 800e3 Aex = 13e-12 alpha = 0.02 m = uniform(1, .1, 0) relax() save(m) // relaxed state autosave(m, 200e-12) tableautosave(10e-12) B_ext = vector(-24.6E-3, 4.3E-3, 0) run(1e-9)
This example should be pretty straight-forward to follow. Space-dependent output is stored in OVF format, which is compatible with OOMMF and can be converted with mumax3-convert. Below is the output converted to PNG.
The data table is stored in a simple text format compatible with gnuplot, like used for the plot below.







Msat = 1000e3
Aex = 10e-12
// define exchange length
lex := sqrt(10e-12 / (0.5 * mu0 * pow(1000e3 ,2)))
d := 30 * lex // we test for d/lex = 30
Sizex := 5*d // magnet size x
Sizey := 1*d
Sizez := 0.1*d
nx := pow(2, ilogb(Sizex / (0.75*lex))) // power-of-two number of cells
ny := pow(2, ilogb(Sizey / (0.75*lex))) // not larger than 0.75 exchange lengths
SetGridSize(nx, ny, 1)
SetCellSize(Sizex/nx, Sizey/ny, Sizez)
m = Uniform(1, 0.1, 0) // initial mag
relax()
save(m) // remanent magnetization
print("<m> for d/lex=30: ", m.average())

SetGridsize(128, 32, 1)
SetCellsize(4e-9, 4e-9, 30e-9)
Msat = 800e3
Aex = 13e-12
m = randomMag()
relax() // high-energy states best minimized by relax()
Bmax := 100.0e-3
Bstep := 1.0e-3
MinimizerStop = 1e-6
TableAdd(B_ext)
for B:=0.0; B<=Bmax; B+=Bstep{
B_ext = vector(B, 0, 0)
minimize() // small changes best minimized by minimize()
tablesave()
}
for B:=Bmax; B>=-Bmax; B-=Bstep{
B_ext = vector(B, 0, 0)
minimize() // small changes best minimized by minimize()
tablesave()
}
for B:=-Bmax; B<=Bmax; B+=Bstep{
B_ext = vector(B, 0, 0)
minimize() // small changes best minimized by minimize()
tablesave()
}
EdgeSmooth=n means n³ samples per cell are used to determine its volume. EdgeSmooth=0 implies a staircase approximation, while EdgeSmooth=8 results in quite accurately resolved edges.
SetGridsize(100, 100, 50)
SetCellsize(1e-6/100, 1e-6/100, 1e-6/50)
EdgeSmooth = 8
setgeom( rect(800e-9, 500e-9) )
saveas(geom, "rect")
setgeom( cylinder(800e-9, inf) )
saveas(geom, "cylinder")
setgeom( circle(200e-9).repeat(300e-9, 400e-9, 0) )
saveas(geom, "circle_repeat")
setgeom( cylinder(800e-9, inf).inverse() )
saveas(geom, "cylinder_inverse")
setgeom( cylinder(800e-9, 600e-9).transl(200e-9, 100e-9, 0) )
saveas(geom, "cylinder_transl")
setgeom( ellipsoid(800e-9, 600e-9, 500e-9) )
saveas(geom, "ellipsoid")
setgeom( cuboid(800e-9, 600e-9, 500e-9) )
saveas(geom, "cuboid")
setgeom( cuboid(800e-9, 600e-9, 500e-9).rotz(-10*pi/180) )
saveas(geom, "cuboid_rotZ")
setgeom( layers(0, 25) )
saveas(geom, "layers")
setgeom( cell(50, 20, 0) )
saveas(geom, "cell")
setgeom( xrange(0, inf) )
saveas(geom, "xrange")
a := cylinder(600e-9, 600e-9).transl(-150e-9, 50e-9, 0 )
b := rect(600e-9, 600e-9).transl(150e-9, -50e-9, 0)
setgeom( a.add(b) )
saveas(geom, "logicAdd")
setgeom( a.sub(b) )
saveas(geom, "logicSub")
setgeom( a.intersect(b) )
saveas(geom, "logicAnd")
setgeom( a.xor(b) )
saveas(geom, "logicXor")
setgeom( imageShape("mask.png") )
saveas(geom, "imageShape")















setgridsize(256, 128, 1)
setcellsize(5e-9, 5e-9, 5e-9)
m = Uniform(1, 1, 0) // no need to normalize length
saveas(m, "uniform")
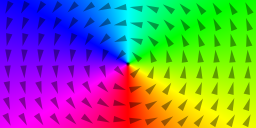
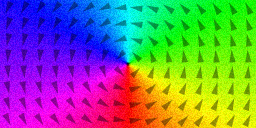
m = Vortex(1, -1) // circulation, polarization
saveas(m, "vortex")
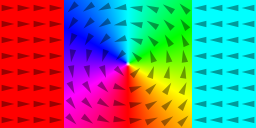
m = TwoDomain(1,0,0, 0,1,0, -1,0,0) // Néel wall
saveas(m, "twodomain")
m = RandomMag()
saveas(m, "randommag")
m = TwoDomain(1,0,0, 0,1,0, -1,0,0).rotz(-pi/4)
saveas(m, "twodomain_rot")
m = VortexWall(1, -1, 1, 1)
saveas(m, "vortexwall")
m = VortexWall(1, -1, 1, 1).scale(1/2, 1, 1)
saveas(m, "vortexwall_scale")
m = Vortex(1,-1).transl(100e-9, 50e-9, 0)
saveas(m, "vortex_transl")
m = Vortex(1,-1).Add(0.1, randomMag())
saveas(m, "vortex_add_random")
m = BlochSkyrmion(1, -1).scale(3,3,1)
saveas(m, "Bloch_skyrmion")
m = NeelSkyrmion(1,-1).scale(3,3,1)
saveas(m, "Néel_skyrmion")
// set m in only a part of space, or a single cell:
m = uniform(1, 1, 1)
m.setInShape(cylinder(400e-9, 100e-9), vortex(1, -1))
m.setCell(20, 10, 0, vector(0.1, 0.1, -0.9)) // set in cell index [20,10,0]
saveas(m, "setInShape_setCell")
//Read m from .ovf file.
m.loadfile("myfile.ovf")
saveas(m, "loadfile")










setgridsize(128, 128, 1)
setcellsize(2e-9, 2e-9, 2e-9)
d := 200e-9
sq := rect(d, d) // square with side d
h := 50e-9
hole := cylinder(h, h) // circle with diameter h
hole1 := hole.transl(100e-9, 0, 0) // translated circle #1
hole2 := hole.transl(0, -50e-9, 0) // translated cricle #2
cheese:= sq.sub(hole1).sub(hole2)// subtract the circles from the square (makes holes).
setgeom(cheese)
msat = 600e3
aex = 12e-13
alpha = 3
// rotate the cheese.
for i:=0; i<=90; i=i+30{
angle := i*pi/180
setgeom(cheese.rotz(angle))
m = uniform(cos(angle), sin(angle), 0)
minimize()
save(m)
}

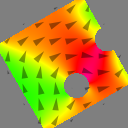


Space-dependent parameters are defined using material regions. Regions are numbered 0-255 and represent different materials. Each cell can belong to only one region. At the start of a simulation all cells have region number 0.
Regions are defined with defregion(number, shape), where shape is explained in the geometry example.
When you're not using regions, like in the above examples, you'll probably set parameters with a simple assign:
Aex = 12e-13Behind the screens, this sets Aex in all regions.
It's always a good idea to output the regions quantity, as well as all your material parameters.
N := 128 setgridsize(N, N, 1) c := 4e-9 setcellsize(c, c, c) // disk with different anisotropy in left and right half setgeom(circle(N*c)) defregion(1, xrange(0, inf)) // left half defregion(2, xrange(-inf, 0)) // right half save(regions) Ku1.setregion(1, .1e6) anisU.setRegion(1, vector(1, 0, 0)) Ku1.setregion(2, .2e6) anisU.setRegion(2, vector(0, 1, 0)) save(Ku1) save(anisU) Msat = 800e3 // sets it everywhere save(Msat) Aex = 12e-13 alpha = 1 m.setRegion(1, uniform(1, 1, 0)) m.setRegion(2, uniform(-1, 1, 0)) saveas(m, "m_inital") run(.1e-9) saveas(m, "m_final")



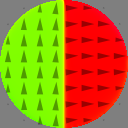


Nx := 256 Ny := 256 Nz := 1 setgridsize(Ny, Nx, Nz) c := 4e-9 setcellsize(c, c, c) setgeom(circle(Nx*c)) Msat = 800e3 Aex = 12e-13 alpha = 1 m = vortex(1, 1) save(m) save(m.Comp(0)) save(Crop(m, 0, Nx/2, 0, Ny/2, 0, Nz)) mx := m.Comp(0) mx_center := CropY(mx, Ny/4, 3*Ny/4) save(mx_center)



Mumax3 has built-in generation of MFM images from the magnetization. The MFM tip lift can be freely chosen. By default the tip magnetization is modeled as a point monopole at the apex. This is sufficient for most situations. Nevertheless, it is also possible to model partially magnetized tips by setting MFMDipole to the magnetized portion of the tip, in meters. E.g., if only the first 20nm of the tip is (vertically) magnetized, set MFMDipole=20e-9.
setgridsize(256, 256, 1) setcellsize(2e-9, 2e-9, 1e-9) setgeom(rect(400e-9, 400e-9)) msat = 600e3 aex = 10e-12 m = vortex(1, 1) relax() save(m) MFMLift = 10e-9 saveas(MFM, "lift_10nm") MFMLift = 40e-9 saveas(MFM, "lift_40nm") MFMLift = 90e-9 saveas(MFM, "lift_90nm")



setGridSize(128, 128, 1) setCellSize(2e-9, 2e-9, 1e-9) Msat = 600e3 Aex = 10e-12 anisU = vector(0, 0, 1) Ku1 = 0.59e6 alpha = 0.02 Xi = 0.2 m = twoDomain(0, 0, 1, 1, 1, 0, 0, 0, -1) // up-down domains with wall between Bloch and Néél type relax() // Set post-step function that centers simulation window on domain wall. ext_centerWall(2) // keep m[2] (= m_z) close to zero // Schedule output autosave(m, 100e-12) // Run for 1ns with current through the sample j = vector(1.5e13, 0, 0) pol = 1 run(.5e-9)






setGridSize(256, 64, 1) setCellSize(3e-9, 3e-9, 10e-9) Msat = 860e3 Aex = 13e-12 Xi = 0.1 alpha = 0.02 m = twodomain(1,0,0, 0,1,0, -1,0,0) notches := rect(15e-9, 15e-9).RotZ(45*pi/180).Repeat(200e-9, 64*3e-9, 0).Transl(0, 32*3e-9, 0) setGeom(notches.inverse()) // Remove surface charges from left (mx=1) and right (mx=-1) sides to mimic infinitely long wire. We have to specify the region (0) at the boundaries. BoundaryRegion := 0 MagLeft := 1 MagRight := -1 ext_rmSurfaceCharge(BoundaryRegion, MagLeft, MagRight) relax() ext_centerWall(0) // keep m[0] (m_x) close to zero // Schedule output autosave(m, 50e-12) tableadd(ext_dwpos) // domain wall position tableautosave(10e-12) // Run the simulation with current through the sample pol = 0.56 J = vector(-10e12, 0, 0) Run(0.5e-9)











ext_makegrains is used to define grain-like regions using Voronoi tessellation. We vary the material parameters in each grain.
N := 256
c := 4e-9
d := 40e-9
setgridsize(N, N, 1)
setcellsize(c, c, d)
setGeom(circle(N*c))
// define grains with region number 0-255
grainSize := 40e-9 // m
randomSeed := 1234567
maxRegion := 255
ext_makegrains(grainSize, maxRegion, randomSeed)
defregion(256, circle(N*c).inverse()) // region 256 is outside, not really needed
alpha = 3
Kc1 = 1000
Aex = 13e-12
Msat = 860e3
// set random parameters per region
for i:=0; i<maxRegion; i++{
// random cubic anisotropy direction
axis1 := vector(randNorm(), randNorm(), randNorm())
helper := vector(randNorm(), randNorm(), randNorm())
axis2 := axis1.cross(helper) // perpendicular to axis1
AnisC1.SetRegion(i, axis1) // axes need not be normalized
AnisC2.SetRegion(i, axis2)
// random 10% anisotropy variation
K := 1e5
Kc1.SetRegion(i, K + randNorm() * 0.1 * K)
}
// reduce exchange coupling between grains by 10%
for i:=0; i<maxRegion; i++{
for j:=i+1; j<maxRegion; j++{
ext_ScaleExchange(i, j, 0.9)
}
}
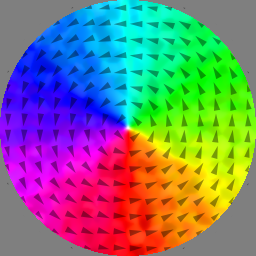
m = vortex(1, 1)
run(.1e-9)
save(regions)
save(Kc1)
save(AnisC1)
save(AnisC2)
save(m)
save(exchCoupling)





scale = (RKKY * cellsize_z) / (2 * Aex).
N := 10
setgridsize(N, N, 2)
c := 1e-9
setcellsize(c, c, c)
defRegion(0, layer(0))
defRegion(1, layer(1))
Msat = 1e6
Aex = 10e-12
RKKY := -1e-3 // 1mJ/m2
scale := (RKKY * c) / (2 * Aex.Average())
ext_scaleExchange(0, 1, scale)
tableAdd(E_total)
m.setRegion(0, uniform(1, 0, 0))
for ang:=0; ang<360; ang++{
m.setRegion(1, uniform(cos(ang*pi/180), sin(ang*pi/180), 0))
t = ang * 1e-9 // output "time" is really angle
tablesave()
}
FixedLayer = ..., which can be space-dependent. The spacer layer properties are modeled by setting the parameters Lambda and EpsilonPrime. Finally Pol sets the current polarization and J the current density, which should be along z in this case.
Below we switch an MRAM bit.
// geometry sizeX := 160e-9 sizeY := 80e-9 sizeZ := 5e-9 Nx := 64 Ny := 32 setgridsize(Nx, Ny, 1) setcellsize(sizeX/Nx, sizeY/Ny, sizeZ) setGeom(ellipse(sizeX, sizeY)) // set up free layer Msat = 800e3 Aex = 13e-12 alpha = 0.01 m = uniform(1, 0, 0) // set up spacer layer parameters lambda = 1 Pol = 0.5669 epsilonprime = 0 // set up fixed layer polarization angle := 20 px := cos(angle * pi/180) py := sin(angle * pi/180) fixedlayer = vector(px, py, 0) // send current Jtot := -0.008 // total current in A area := sizeX*sizeY*pi/4 jc := Jtot / area // current density in A/m2 J = vector(0, 0, jc) // schedule output & run autosave(m, 100e-12) tableautosave(10e-12) run(1e-9)











Shift function, we can shift the system (magnetization, regions and geometry) by a given number of cells. Here we use this feature to simulate a moving hard disk platter. A time-dependent gaussian field profile mimics the write field.
Nx := 512
Ny := 128
c := 5e-9
setgridsize(Nx, Ny, 1)
setcellsize(c, c, c)
ext_makegrains(30e-9, 256, 0)
// PMA material
Ku1 = 0.4e6
Aex = 10e-12
Msat = 600e3
alpha = 1
delta := 0.2 // anisotropy variation
for i:=0; i<256; i++{
// random cubic anisotropy direction
AnisU.SetRegion(i, vector(delta*(rand()-0.5), delta*(rand()-0.5), 1))
// strongly reduce exchange coupling between grains
for j:=i+1; j<256; j++{
ext_scaleExchange(i, j, 0.1)
}
}
m = uniform(0, 0, 1)
// Gaussian external field profile
mask := newVectorMask(Nx, Ny, 1)
for i:=0; i<Nx; i++{
for j:=0; j<Ny; j++{
r := index2coord(i, j, 0)
x := r.X()
y := r.Y()
Bz := exp(-pow((x-500e-9)/100e-9, 2)) * exp(-pow(y/250e-9, 2))
mask.setVector(i, j, 0, vector(0, 0, Bz))
}
}
// 500Mbit/s oscillating write field
f := 0.5e9
A := 1.5
B_ext.add(mask, -A*sin(2*pi*f*t))
autosave(m, 600e-12)
// Spin the hard disk
ShiftMagR = vector(0, 0, 1) // new magnetization to enter
for i:=0; i<120; i++{
run(30e-12)
Shift(-1) // one cell to the left
}






